Do people really
get bright ideas in the shower? That is a topic for another day, but I think I
recently had a good one. It all started with a question from a post
baccalaureate trainee at NIH, where I currently work. She encountered a problem
when designing primers for Polymerase Chain Reaction (PCR) based on a published
paper. Since I was the resident PCR specialist she came to me for advice.
I think a brief
introduction to PCR will help here, though a detailed blog on PCR will follow. It
is the biochemical reaction that revolutionized DNA amplification, the basis
for genome sequencing and cloning. It involves choosing two short stretches of
DNA (primers) from a template (genome, plasmid, etc) and exponentially amplifying
the DNA between primers using an enzyme called DNA polymerase. DNA, such as
our genome, consists of two complementary strands running in opposite
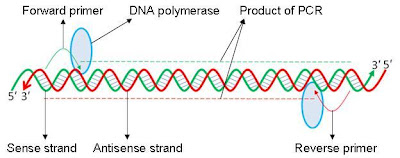
directions, while PCR primers are single-stranded. For PCR to work, each chosen
primer needs to bind one template strand and the two primers need to be
directed towards each other (illustrated below).
The postbac’s problem
was that of the two published primer sequences, only one could be located in
the mouse genome. Furthermore, she was surprised that what was labeled as
forward primer in the paper was actually a reverse primer! By convention, the
primer that is identical to the sequence of your gene of interest is called
forward primer and the one that resembles the complementary strand is called
the reverse primer. The postbac was confused because the paper labeled the
primer from the complementary strand as forward. But does that really matter? As
an aside, this actually happens with surprising frequency; scientists don’t
seem to have enough time to confirm many of their findings and rush their
publications out the door. Naturally once a finding is out, very few try to
repeat those experiments; what is the point anyway… if you confirm the results,
then you would have wasted your time and resources on something that is already
known!
Getting back to
PCR, the primers don’t really know whether they are forward or reverse; if they
see DNA sequence that they can bind to, they will. What we name the primers is
up to us. They don’t care what we call them, forward/reverse, left/right,
up/down, or mango/banana. Put them in the PCR reaction, they work the same
whatever their names.
So, what was my
shower idea? I happened to be thinking about my drive through Highway US 163, a two-lane road with one lane going in
each direction, near Monument
Valley in Utah/Arizona a
few years ago. This is where Forrest Gump stopped running. He probably did not
know which direction he was running in, but he knew how to run! Seemed like a
great locale to illustrate how PCR works... Now drop a couple of drivers on
that straight road and tell them whichever direction they go is the right one.
The only way they see each other is if they start driving in opposing
directions and are heading towards each other, assuming they drive at the same
speed. It is immaterial who drives towards Utah
and who drives towards Arizona.
The objective is for them to meet. This is what needs to happen for PCR to
work. What are the parallels to PCR? The two opposing lanes of US 163 are the
two strands of DNA, the two cars are the primers and the drivers are the
polymerase enzymes. So, the bottom line for my postbac friend was, not to worry
about the nomenclature, but to design the primers and see if they work!
In this illustration, the thick
green line corresponds to the sense strand (the gene sequence) and the thick
red line corresponds to the antisense strand. Thin green line is the
forward/sense primer and thin red line is the reverse/antisense primer. Blue
oval is the DNA polymerase enzyme. PCR product would be a double helix formed by the two
dashed lines (red and green). The DNA molecules have arrows because DNA is
directional. 5′ represents the end of
DNA that has a Phosphate group and 3′
represents the end of DNA with the sugar group. In this example, a green sequence will always bind a red sequence and vice versa.
No comments:
Post a Comment